This web page was produced as an assignment for Genetics 677, an undergraduate course in UW Madison
The complexity of GSK3B and GO
The three major sources of information we can find in Gene Ontology website [27] can answer 3 important questions about the molecule:1st –the biological processes it is involved in, 2nd- the cellular components it is found in or even temporary complexes it forms with other molecules, and 3rd – the molecular functions assessed to it. According to it the general characteristics of GSK3B are as follows:
Gene Ontology:
1.Biological Processes: ER overload response, Wnt receptor signaling pathway through beta-catenin , glycogen metabolic process , peptidyl-serine phosphorylation, positive regulation of protein complex assembly, positive regulation of protein export from nucleus
2. Cellular component: Axin-APC-beta-catenin-GSK3B complex, beta-catenin destruction complex, cytosol , nucleus .
3. Molecular function: ATP binding, NF-kappaB binding, beta-catenin binding, glycogen synthase kinase 3 activity, p53 binding, protein kinase A catalytic subunit binding, tau-protein kinase activity.
(Data retrieved from UniProt: http://www.uniprot.org)
In order to understand the specific mechanism by which this kinase interact with other biological molecules and the mechanisms by which it regulates key cellular processes we need to understand the details of the enzyme’s primary, secondary and tertiary structure as well as the amino acid residues involved in this interaction. Many of these amino acids are subjected to additional post-translational modifications such as phosphorylation and can alter the interactive ability of the molecule or even its function in general. By using variety of web-based tools and programs such as Prosite, UniProt, Expasy and comparing the predicted bioinformatic data with the experimental ones, from the published literature, I am going to create a brief overview of what is known about the function of GSK3B and how genetic and chemical manipulations can cause help us understand the major processes in which it is involved.
Structure and Function in general.
All three web-based programs (SMART, PROSITE, UNIPROT) identify a key Serine/ Threonine Kinase domain in the structure of the GSK3B. This domain is found in variety of species and seems to be highly conserved. For my analysis I will use data obtained primarily from UniProt due to the fact that the information there is well organized and also allows a quick access to additional programs for reference.
Primary Structure GSK3B
Serine/Threonine kinase domain.
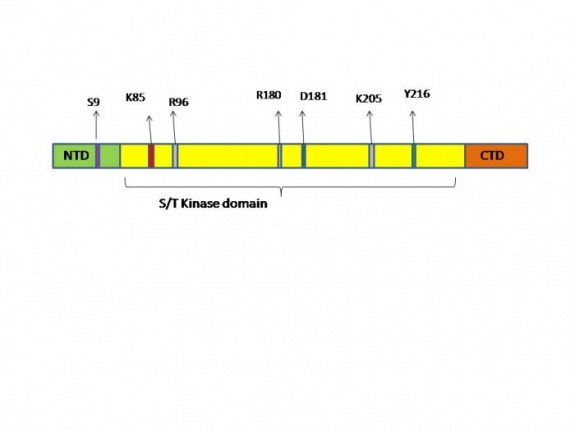
The total length of the molecule consists of 420 amino acid residues and the Kinase domain is identified to be between 56-340 amino acid residues:
MSGRPRTTSF AESCKPVQQP SAFGSMKVSR DKDGSKVTTV VATPGQGPDR PQEVSYTDTK
VIGNGSFGVV YQAKLCDSGE LVAIKKVLQD KRFKNRELQI MRKLDHCNIV RLRYFFYSSG
EKKDEVYLNL VLDYVPETVY RVARHYSRAK QTLPVIYVKL YMYQLFRSLA YIHSFGICHR
DIKPQNLLLD PDTAVLKLCD FGSAKQLVRG EPNVSYICSR YYRAPELIFG ATDYTSSIDV
WSAGCVLAEL LLGQPIFPGD SGVDQLVEII KVLGTPTREQ IREMNPNYTE FKFPQIKAHP
WTKVFRPRTP PEAIALCSRL LEYTPTARLT PLEACAHSFF DELRDPNVKL PNGRDTPALF
In this region a specific Lysine residue (K 85) is assessed to bind ATP. Even though both PROSITE and UniProt determine this residue as a binding site for the ATP there is slight differences of the Nucleotide binding domain that they determine. According to UniProt the Nucleotide binding region spreads between 62-70 amino acid residues (the region in green)while PROSITE extends this region all the way to the 86th amino acid (green and light blue) based on a consensus pattern ([LIV] - G - {P} - G - {P} - [FYWMGSTNH] - [SGA] - {PW} - [LIVCAT] - {PD} - x - [GSTACLIVMFY] - x(5,18) - [LIVMFYWCSTAR] - [AIVP] - [LIVMFAGCKR] - K ;K binds ATP.). This data is further supported by genetic experiments in Arabidopsis thaliana[21] and Xenopus[22], where the substitution of the Lysine residue to Argenine (K85→R) creates “dead” enzyme – the molecule cannot bind ATP and becomes inactive.
D181 Active site.
As the active site of the kinase both sites determine the aspartic acid on position 181 (D181 )to play this role and even specify its function (UniProt) as a proton acceptor. Interestingly I was not able to find any experimental studies done with this residues and the effect of possible substitution on biological processes. My assumption is that the defects that will be observed are going to be similar to the K85R substitutions, due the fact that a “dead” enzyme will be created.
S9 – auto inhibition.
Another very interesting residue that we need to mention is the Serine amino acid in position 9 (S9). This site is known to be phosphorylated by variety of kinases one of which, PKB (also known as Akt), was largely studied. The effect of the phosphorylation has been proven to inhibit the
Y216 – increases the activity.
Some protein kinases closely related to GSK3 (CDK2, p38γ and ERK2) are known to be activated after a specific residue is phosphorylated on their activation loop (also known as T-loop)[19]. For some (ERK2 and p38γ) phosphorylation of a tyrosine residue on the T-loop leads to opening of the catalytic side of the enzyme [19]. In GSK3 Tyr in position 216 (Y216) plays the role of activator of the molecule. Scientists believe that the phosphorylation of the Y216 facilitate subsequent substrate phosphorylation but is not strictly required for it, due to the fact that the T-loop in GSK3B does not prevent substrate binding [23]. Therefore phosphorylation of Y216 might have evolved as a mechanism for a fine “tuning” of the activity of this kinase.
R96, R180 and K205 – pocket for binding of primed substrates.
The last set of residues that I need to discuss before proceeding with the mechanism of action are three positively charged residues Arg 96, Arg 180 and Lys 205 (R96, R180 and K205). These residues form a pocket in the 3D structure of the protein and help primed substrates bind with specific orientation to the GSK3B. The primed substrates for GSK3 have a conserved sequence Ser-P/Thr-P – X – X – X – Ser/Thr (X- are any kind of amino acids). In this sequence the first S/T residue is phosphorylated by a different kinase, which increases the affinity of the molecule to the GSK3B, this increase of affinity is due to the positive pocket in the GSK3B which help orient the substrate in a position where the S/T residue can be phosphorylated from the GSK3B [19, 23](Figure 1A). The importance of this positively charged pocket and especially the role of R96 has been proven experimentally where the R96 is substituted for Alanine (R96→A) and reduces significantly the affinity of the GSK3B to primed substrates [21](Figure 1B).
Regulation of the enzymatic activity
Figure 1A [19] explains clearly a proposed mechanism by which GSK3B interacts with its substrates. GSK3B is known to be constitutively active and can phosphorylate both primed and unprimed substrates. When S9 residue, on the N-terminal domain, is phosphorylated by kinases regulators of GSK3B, this domain becomes a pseudosubstrate, in other words it mimics a primed substrate and can bind to the positively charged pocket of the kinase domain, preventing it from binding to other substrates. This autoinhibitory mechanism has been studied extensively [19, 21,23] and is also supported by genetic experiments [19] where substitution of S9 to Alanine – leads to loss of self-inhibition of the molecule and creates specific phenotypes [23]. Phosphorylation of the S9 is known to be performed by PKB/Akt but also other stimuli can lead to the same affect – such as: EG and PDGF that stimulate MAPKAP-K1 (which is GSK3-inactivating kinase); amino acid stimuli which activate p70 ribosomal S6 kinase; activators of cAMP- activated protein kinase (PKA) and activators of PKC.
Regulation of glycogen synthase kinase 3
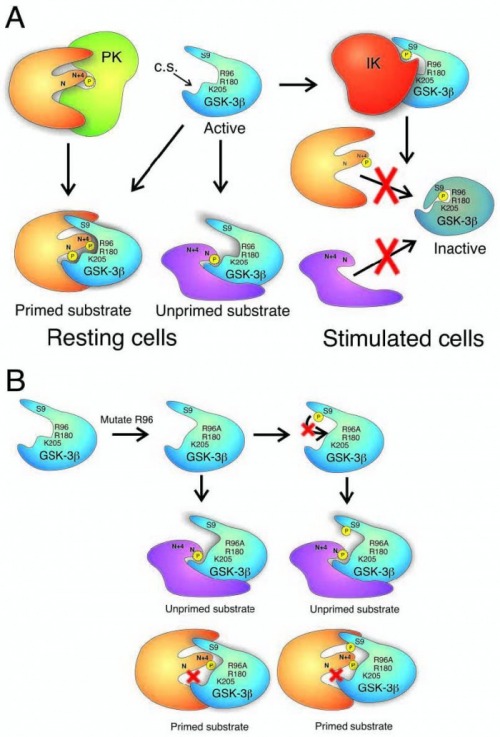
Fig. 1. (A) Regulation of GSK-3b activity by serine phosphorylation. In the resting cell, GSK-3b is constitutively active. Both unprimed substrates and substrates phosphorylated by a priming kinase (PK) are capable of being phosphorylated by the active GSK-3b. The priming phospho-residue at position N + 4, binds a pocket of positive charge arising from the arginine (R) and lysine (K) residues indicated. This directs a serine or threonine at position N to the active catalytic site (C.S.). When an inactivating kinase (IK) such as PKB/Akt phosphorylates GSK-3b on serine 9 (S9), the phosphorylated Nterminus becomes a primed pseudo-substrate that occupies the positive binding pocket and active site of the enzyme, acting as a competitive inhibitor for true substrates. This prevents phosphorylation of any substrates. (B) Effect of mutating arginine 96 to alanine (R96A) on GSK-3b activity. Since arginine 96 is a crucial component of the positive pocket that binds primed substrates, its mutation to an
uncharged alanine residue disrupts the pocket so that primed substrates can no longer bind. The enzyme retains activity. Also, the S9-phosphorylated pseudosubstrate is no longer capable of inactivating the enzyme. As a consequence, GSK- 3b, whether S9-phosphorylated or not, can phosphorylate unprimed substrates, but not primed substrates. Note that unprimed and primed substrates interact with GSK-3 through different interfaces.
Doble, B. & Woodgett, J. (2003). GSK-3: tricks of the trade for multi-tasking kinase. Journal of cell science 116, 1175-1186
GSK3B and Wnt canonical pathway.
Phosphorylation of the S9 residue is not the only way by which GSK3B is regulated in the cells. GSK3B is part of very important component in the Wnt/canonical pathway. In resting cells the enzyme is believed to be a central part of a protein complex (Figure 2) [19] which involves the following molecules: GSK3B, APC- a tumor suppressor protein and Axin – which serves as a scaffold protein and has many protein-protein interaction domains. Both Axin and APC are phosphorylated by GSK3B which increases the stability of the complex and the binding of β-catenin to it. Β-catenin is known to be a primed substrate for GSK3B. CK1 is known to phosphorylate β-catenin on the S45 residue. In addition this kinase binds and also phosphorylates axin, Dishevelled and APC. After β-catenin is phosphorylated by GSK3B it becomes target for ubiquitylation and subsequent degradation by the proteasome. Stimulation of the Wnt pathway and via the Frizzled receptor causes the “destructive” complex to disassemble, which stops the phosphorylation and degradation of the β-catenin and leads to its accumulation in the cytoplasm and later translocation in the nucleus. In the nucleus β-catenin activates transcription of specific genes by binding to the LEF/TCF transcription factor.
GSK3B and Wnt canonical pathway
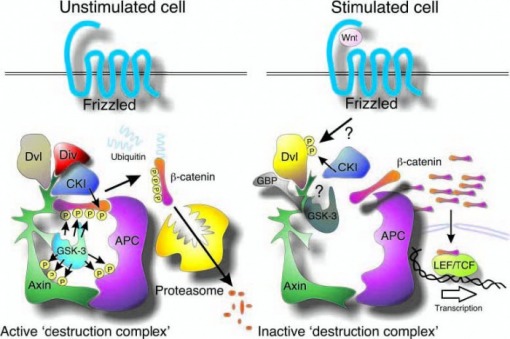
Fig. 2. Central role of GSK-3 in the Wnt/b-catenin pathway. In unstimulated cells, CKI phosphorylates b-catenin on S45, priming it for subsequent phosphorylation by GSK-3 (S41, S37, S33), which targets b-catenin for ubiquitylation and proteasomal degradation. The ankyrin repeat protein, Diversin (Div), may help recruit CKI to the destruction complex. Wnt stimulation activates the receptor Frizzled, which then signals through Dishevelled (Dvl), using an unclear mechanism, to inactivate b-catenin phosphorylation. Unphosphorylated b-catenin accumulates and then translocates to the nucleus where it transactivates genes regulated by TCF/LEF transcription factors. The GSK-3- binding protein (GBP/FRAT) may be involved in transmission of a Wnt signal by regulating binding of GSK-3 to the scaffold protein, axin.
Doble, B. & Woodgett, J. (2003). GSK-3: tricks of the trade for multi-tasking kinase. Journal of cell science 116, 1175-1186
Summary:
The mechanisms by which GSK3B is regulated and perform its regulatory functions are very distinct to each other and reveal the complexity of the molecule. This enzyme is highly conserved in variety of species and it will be interesting to observe the genetic manipulations that were employed in some studies and the general phenotypic results obtained from them. I have examined a few studies done with mice[2], Zebrafish [20], Xenopus [22], and Arabidopsis thaliana [21] to analyze and compare the physiological processes in which GSK3B is involved as well as the differences observed in the different organisms. These differences can provide us with valuable information of how the enzyme has functionally diverged throughout the evolution and what kinds of functions have been preserved in the organisms. For additional information about the phenotypes please click Here.
Back to: Home page, Phenotypes.
Schematic presentation of the protein primary structure
| Schematic presentation of the secondary structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
Blue – αhelix
Green –β strand (or sheet)
Yellow – turnin the structure
Imageretrieved from UniProt – please refer to the websitefor detailed description of the amino acid residues involved in the formationof these structures
3D Structure of the GSK3B
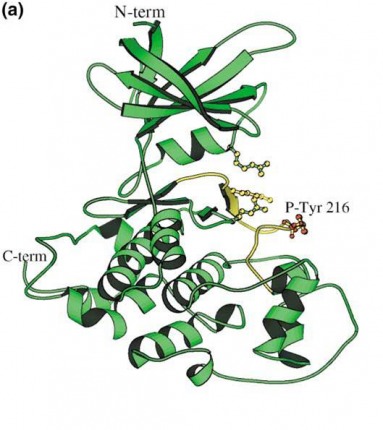
Structure of crystal form A of GSK-3-beta. Phosphotyrosine 216 (red atoms) is labeled. The activation loop is in yellow, and the side chainsof Arg 96, Arg 180, and Lys 205 are shown (yellow atoms).
Bax,B.(December 2001).The structure of phosphorylated GSK-3 beta complexed with a peptide, FRATtide, that Inhibits beta-catenin phosphorylation. Structure. 9, 1143-1152
Trypsin cutting sites
Before an MS analysis is performed on a protein often times it is helpful to digest it with variety of enzymes. One of the most commonly used for this purpose is trypsin. ExPASy website provides a helpful tool (PeptideCuttter) which when provided with amino acid sequence can predict the cutting sites for variety of enzymes. For the GSK3B protein the following positions were identified as targets for trypsin digestion.
| No. of cleavages | Position of the cleavage sites |
| 45 | 6 27 30 36 60 74 85 86 91 92 94 96 102 103 111 113 122 123 141 144 148 150 159 167 180 197 205 209 220 223 271 278 282 292 297 303 316 321 332 341 357 362 367 396 418 |
Theoretical pI/Mw of GSK3B
Another helpful tool offered by ExPASy is Compute pI/Mw – it allows the user to calculate the theoretical isoelectric point and molecular weight of a protein or peptide of interest. This information is useful to identify and predict where the protein/peptide might be found in a 2D SDS-PAGE gel. For the GSK3B amino acid sequence the predicted information is the following:
Theoretical pI = 8.98 ; Mw = 48033.67
It is also important to remember that post translational modifications of the protein such as phosphorylation for example can change these values.
Subcellular localization
There are many web-based software programs for prediction of the subcellular localization of the protein sequences. Unfortunately even after using the same sequence for the GSK3B protein the results between the programs showed significant deviations. Some specific examples that could be demonstrated are as follows:
Proteome analyst - predicted the cytoplasm with 100% confidence level.
pTarget – determined the nuclear localization of the protein with 81.4% confidence
TargetP – identifies mitochondrial location but the reliability of the result was rated 5 (which is the lowest for this website)
PSORTII – is software I personally found most informative. It not only creates a detailed list of the sub-programs that are used for identification of the subcellular localization but also provides a link for each specific sub-program with comments about the methods and algorithms used and general reliability of these tools. The cumulative results obtained from the program are based on k-NN prediction (k-nearest neighbor algorithm) and are the following:
k = 9/23
52.2 %: nuclear
26.1 %: cytoplasmic
13.0 %: mitochondrial
8.7 %: cytoskeletal
>> prediction for QUERY is nuc (k=23)
Conclusion: based on the inconsistencies of the results obtained from the web-based prediction methods the data obtained from them need to be cautiously addressed. At this moment experimental procedures remain the most reliable source of evidence for determining the cellular localization of the proteins.
References
2. Kerkela, R. (2008). Deletion of GSK-3 beta in mice leads to hypertrophic cardiomyopathy secondary to cardiomyoblast hyperproliferation. The journal of clinical investigation, 118, iss:11, 3609 -3618
19.Doble, B. & Woodgett, J. (2003). GSK-3: tricks of the trade for multi-tasking kinase. Journal of cell science 116, 1175-1186
20.Lee, H. & Tsai, J. (August, 3 2007). Glycogen synthase kinase 3α and 3β have distinct functioning during cardiogenesis of Zebrafish embryo. BMC developmental biology, 7:93.
21. Claisse, G. & Charrier, B. (February, 15 2007). The Arabidopsis thaliana GSK3/Shaggy like kinase AtSK3-2 modulates floral cell expansion. Plant Mol Biol 64:113-124.
22. Pierce, S. & Kimelman, D. (1995). Regulation of Spemann organizer formation by the intracellular kinase Xgsk-3. Development 121, 755-765.
23. Dajani,R. (2001). Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105, 721-732.
24. PROSITE: http://www.expasy.ch/prosite
25. ExPASy: http://www.expasy.ch
26. UniProt: http://www.uniprot.org
27. Gene Ontology: http://www.ebi.ac.uk/GO
28. Bax,B.(December 2001).The structure of phosphorylated GSK-3 beta complexed with a peptide, FRATtide, that Inhibits beta-catenin phosphorylation. Structure. 9, 1143-1152
29. Proteome analyst: http://pasub.cs.ualberta.ca:8080/pa/Subcellular
30. pTarget: http://bioapps.rit.albany.edu/pTARGET/
31. TargetP: http://www.cbs.dtu.dk/services/TargetP/
32.PSORTII: http://www.psort.org/
Back to Home page
Contact information
This webpage was created by: Eva Dimitrova, [email protected]
