This web page was produced as an assignment for Genetics 677, an undergraduate course in UW-Madison
Homology in the protein domains
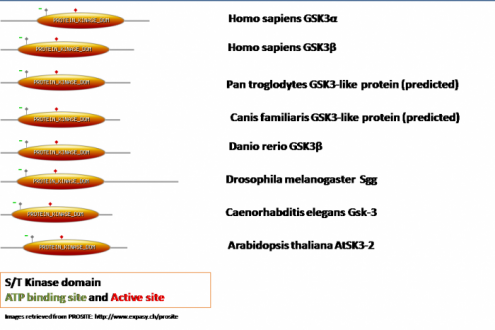
All three web-based programs (SMART, PROSITE, UNIPROT) identify a key Serine/ Threonine Kinase domain in the structure of the GSK3B. This domain is found in variety of species and seems to be highly conserved. For my analysis I will use data obtained primarily from Prosite to demonstrate the common elements between the homologues of GSK3B.
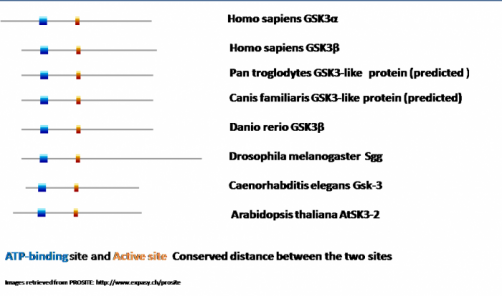
As we can see in all homologues the key serine/threonine kinase domain is present and its relative size is highly conserved. The main difference between the proteins comes from the length of the C- or N- terminal domain. In all of them there is an ATP-binding site (presented in green) and an active site (presented in a red square). In order to determine the relative level of conservation of these two sitesI aligned these sites again (see diagram bellow).
Analysis
What was noticeable from the results is that regardless of the species and the molecule as a whole, the distance between the active site and the ATP-binding domain is highly conserved. This might have to do with the fact that in order the kinase to phosphorylate its substrates it has to use the energy obtained from hydrolysis of ATP. This important function has not been modified in any of the homologues throughout the evolution and this is the optimal distance between the two sites in order for phosphorylation to occur. From most of the research literature published I was able to find experiments that were conducted by modifying the ATP-binding site. Changes of the amino acid residue there cause loss of function mutations and inability of the kinase to phosphorylate its substrates. It will be interesting to design a study where the distance between the two sites is altered and observe how these changes affect the overall performance of the protein.
Back to Home page
What was noticeable from the results is that regardless of the species and the molecule as a whole, the distance between the active site and the ATP-binding domain is highly conserved. This might have to do with the fact that in order the kinase to phosphorylate its substrates it has to use the energy obtained from hydrolysis of ATP. This important function has not been modified in any of the homologues throughout the evolution and this is the optimal distance between the two sites in order for phosphorylation to occur. From most of the research literature published I was able to find experiments that were conducted by modifying the ATP-binding site. Changes of the amino acid residue there cause loss of function mutations and inability of the kinase to phosphorylate its substrates. It will be interesting to design a study where the distance between the two sites is altered and observe how these changes affect the overall performance of the protein.
Back to Home page
References
1.PROSITE: http://www.expasy.ch/prosite
2. ExPASy: http://www.expasy.ch
3. UniProt: http://www.uniprot.org
4. SMART: http://smart.embl-heidelberg.de
2. ExPASy: http://www.expasy.ch
3. UniProt: http://www.uniprot.org
4. SMART: http://smart.embl-heidelberg.de