This web page was produced as an assignment forGenetics 677, an undergraduate course in UW Madison
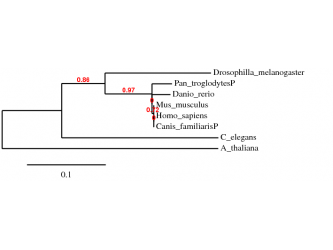
Phylogeny T-coffee alignment
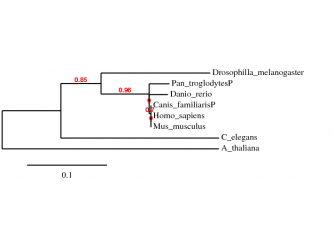
Phylogeny ClustalW
Method: using the “A la Carte” mode from Phylogeny.fr T-coffee alignment was selected, followed by Gblocks alignment curation and visualized with TreeDyn. The convenience of this website allows the building of the tree to be accomplished in one step after customizing the settings
Method: using the “A la Carte” mode from Phylogeny.fr ClustalW alignment was selected, followed by Gblocks alignment curation and visualized with TreeDyn. The convenience of this website allows the building of the tree to be accomplished in one step after customizing the settings.
Comments
As we can see the results obtained from the two alignments are almost the same – an interesting observation is the position of the chimpanzee on the phylogenic trees which could be explained by the fact that the provided protein sequence that was used for the alignment was predicted and not complete. In order to confirm the results the procedure was repeated by using the GeneBee TreeTop building program.
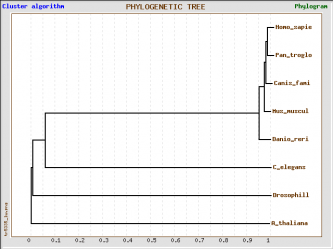
TreeTop T-coffee alignment
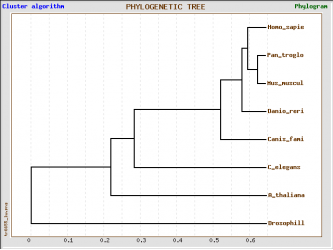
TreeTop ClustalW alignment
Method:The raw data was aligned using T-coffee (which is Mlalign_id_pair algorythm), the aligned data was converted in Phylis format from the Phylogeny.fr website and used to build a tree from TreeTop website using the default settings
Method: the raw data was aligned using ClustalW algorithm, the aligned data was converted into Phylis format from the Phylogeny fr. Website and used to build a tree from TreeTop website using the default settings
Comments
There are not significant differences between the alignment using the two algorithms and two programs. All trees reveal close relations between humans and mice, dog, zebrafish and chimpanzee, which means that the last four species could be used as model organisms to design experiments that can help us understand the mechanism by which GSK3B functions and possible effects that inhibition or activation of the kinase might have in humans.
Back to Home page
References
1.Phylogeny fr: http://www.phylogeny.fr
2.Gene Bee: http://www.genebee.msu.su
Contact information
This webpage was created by: Eva Dimitrova, [email protected]